DNA Sequencing
Second-generation DNA sequencing is transforming the life sciences. This technology, remarkably, offers several orders of magnitude increase in data output and a dramatic reduction in cost compared with conventional sequencing. It enables whole genome sequencing (WGS), targeted resequencing (e.g.) of individual exomes, and custom regions of interest, gene expression and regulation, small RNA discovery, methylation profiling, and genome-wide protein-DNA interaction analysis, in the one platform.
Our facility is managed by specialist molecular biologists/geneticists who assist with experiment design and implementation, library preparation, and troubleshooting. We develop new applications and methodologies in 2GS and optimize workflows (e.g.) comparing whole-exome capture platforms and reduce costs by pre-capture pooling (manuscript in preparation), and work with researchers.
QBI was the first research institute in Australia to acquire the advanced technology platform HiSeq 2000 DNA sequencer from Illumina, capable of delivering 32Gb of DNA sequence per day. This state-of-the-art genomics technology platform delivers QBI researchers working across the species spectrum (C. elegans, fruit fly, honeybee, zebrafish, rodent and human) and applications spectrum (WGS, RNAseq, chIPseq, targeted resequencing including whole exome and custom capture) previously unavailable genetic information that will dramatically enhance research programs in neuroscience.
Instrumentation
Second Generation DNA Sequencing (2GS)

HiSeq 2000 Specifications
Sequence Output:
| Optimal Cluster Density (per mm2) | Per Lane1 | Per FC | Run Time (100bp PE) | |
| Version 2 (v.2) Flow cell | 400-450K/mm2 | 18Gb | 144Gb | 8 days |
| Version 3 (v.3) Flow cell | 750-850K/mm2 | 40Gb | 320Gb | 10 days |
2 flow cell positions (A & B), can be run simultaneously and independently
- We run v.2 and v.3 flow cells
- 8 lanes per flow cell
- Multiplexing up to 48 samples per lane (using unique 6bp molecular barcodes)
TruSeq DNA Sample Prep: 24 different barcode sequences. We run up to 48 different barcodes in a single lane.
Samples are prepared for sequencing by randomly fragmented DNA, to which universal adaptors are ligated, prior to linear amplification of DNA molecules on the flow cell surface. Samples prepared for DNA sequencing are referred to as ‘libraries’, whether starting from total RNA, genomic DNA, selected regions of genomic DNA (targeted resequencing), and more. Protocols for generating DNA libraries differ at the ‘front-end’, depending on the starting nucleic acid (RNA, DNA) and the application, but ultimately double-stranded, adapter-ligated DNA short DNA fragments form what is known as the “DNA library”.
Clonal amplification DNA molecules in the library results in “clusters” of molecules derived from original adapter-ligated random fragments. Cluster generation takes place automatically using the cluster robot (c-Bot). Flow cells are then subject to Sequence By Synthesis (SBS) on the HiSeq 2000.
Applications/Techniques Run at QBI
- Whole Genome Sequencing (WGS)
- Whole transcriptome sequencing (RNAseq)
- Digital gene Expression (RNAseq)
- Targeted resequencing (using in-solution hybridization capture)
- Whole-exome sequencing (NimbleGen SeqCap EZ exome, Illumina TruSeq, and Agilent SureSelect)
- MeDIPseq
- Development methods
Service provision/External samples
QBI’s genomics facility is primarily for the use by QBI researchers. However, the facility will accept libraries from external researchers by prior arrangement. External researchers must provide pre-prepared DNA libraries, which will be subject to quality control by the facility. We do not offer library preparation as a service. Our facility operates on a purely cost recovery basis.
Instrumentation for quality control of DNA library preparation
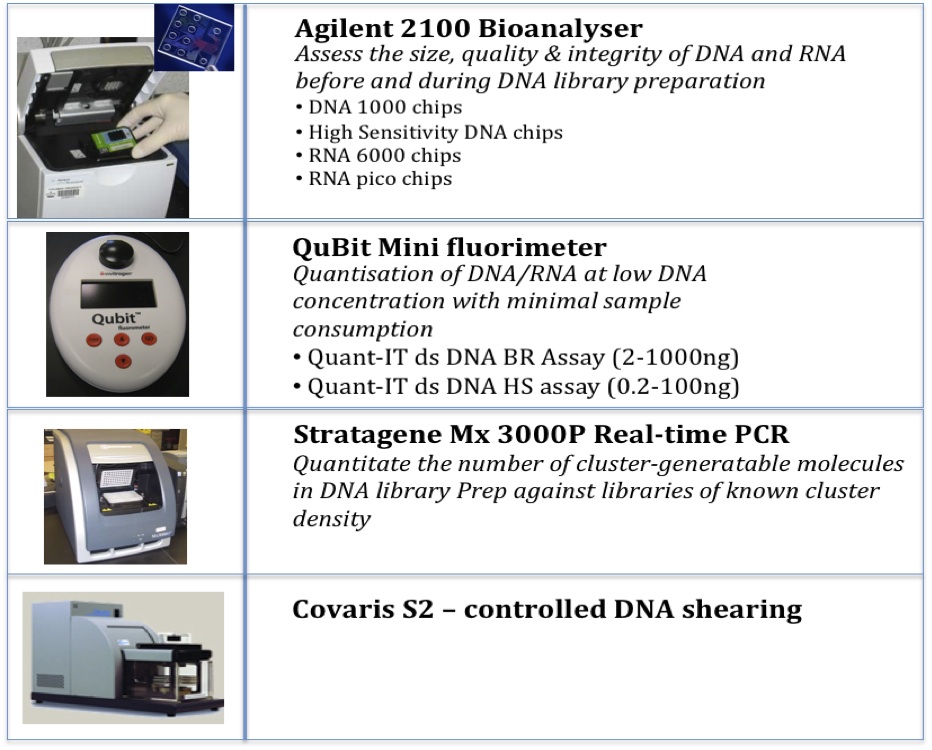
Instrumentation to aid in the preparation of high quality DNA libraries for multiple applications
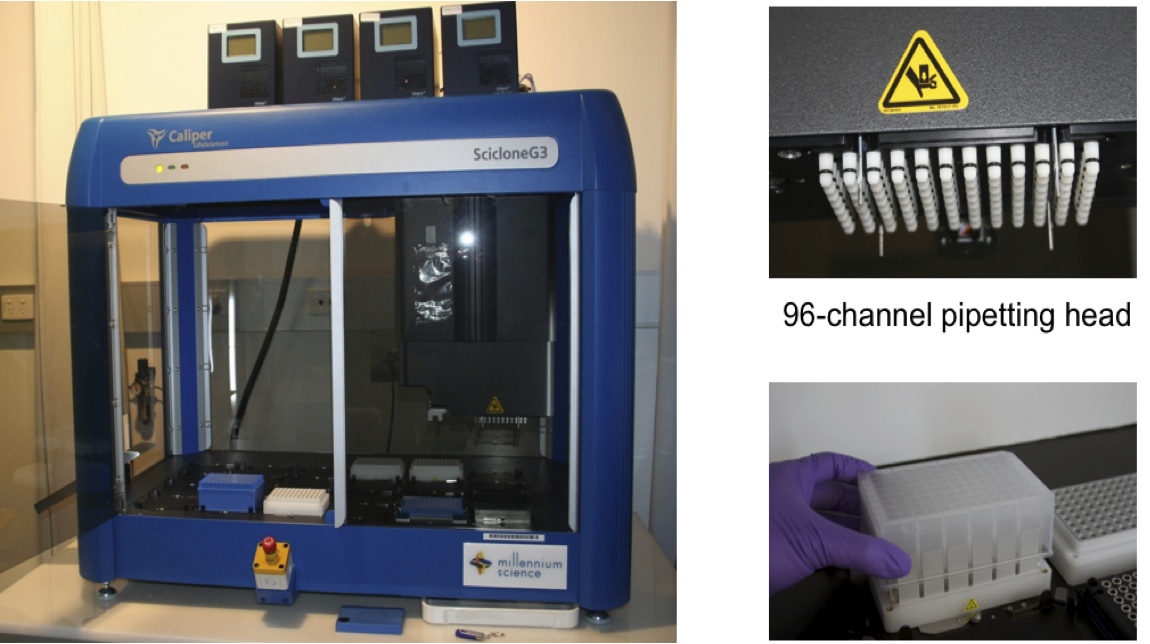
Caliper SciClone G3 Automated DNA library preparation allows us to undertake large sample number projects (e.g.) 1,000 whole-exome captures, enabling variant detection in disease state with high statistical significance (e.g.) in schizophrenia (SZ), ADHD, autism. We have validated protocols for the preparation of Illumina TruSeq DNA libraries and NimbleGen whole exome or custom sequence capture libraries, and we are internally validating protocols for RNAseq library preparation.
PippinPrep (50bp-1.5kb) | Sage BioScience
Semi-automated DNA size selection for Next Generation DNA Sequencing libraries, ensuring high recovery of DNA, ‘tight’ size distribution of fragments, for more diverse, higher coverage libraries.
Other Genomics Platforms in QBI’s Genomics Facility (validation)
Sequenom MassArray 4


